<!-- TODO add slide numbers & maybe slide name --> ### Community Detection and Model Selection in SBMs  Eric W. Bridgeford | {Biostatistics, BME, CIS}<br> [ericwb95@gmail.com](mailto:ericwb95 at gmail dot com) | <https://neurodata.io/talks/commdetect.html> <!-- <br><br><br> --> <!-- <img src="images/logo_jhu.png" STYLE="HEIGHT:50px;"/> --> --- name:talk ### Outline - [Background](#back) - [SBMs](#sbm) - [Community Detection](#commdetect) - [Model Selection](#modselect) --- name:back ### Outline - Background - [SBMs](#sbm) - [Community Detection](#commdetect) - [Model Selection](#modselect) --- ### Background readings - [Chapter 4 -- Network basics](https://docs.neurodata.io/graph-stats-book/representations/ch4/ch4.html) - [Chapter 5.2 -- SBMs](https://docs.neurodata.io/graph-stats-book/representations/ch5/single-network-models_SBM.html) - [Chapter 5.3 -- RDPGs](https://docs.neurodata.io/graph-stats-book/representations/ch5/single-network-models_RDPG.html) - [Chapter 6.1 -- SBM estimation](https://docs.neurodata.io/graph-stats-book/representations/ch6/estimating-parameters_mle.html) - [Chapter 6.4 -- RDPG estimation](https://docs.neurodata.io/graph-stats-book/representations/ch6/estimating-parameters_spectral.html) --- name:sbm ### Outline - [Background](#back) - SBMs - [Community Detection](#commdetect) - [Model Selection](#modselect) --- ### SBM - The nodes will represent regions of the brain (70) - The edges will represent whether two regions are connected or not by a diffusion fiber tract - The community membership vector $\vec z$ indicate which hemisphere (R/L) each node is in - Block matrix $B$ has entries of $0.5$ for the L/L and R/R entries - $0.2$ for the R/L and L/R entries --- ### Defining the parameters of an SBM ```python # import graspologic package import graspologic as gp ``` ```python # set the number of nodes per hemisphere nl = 35; nr = 35 ns = [nl, nr] # the block matrix B = [[0.5, 0.2], [0.2, 0.5]] # the block matrix # the hemisphere of each node zvec = ["L" for i in range(0, nl)] + ["R" for i in range(0, nr)] # one-hot-encode node communities zvec_ohe = np.vstack([[1,0] for i in range(0, nl)] + [[0, 1] for i in range(0, nr)]) # the probability matrix P = zvec_ohe @ B @ zvec_ohe.transpose() ``` ```python fig, axs = plt.subplots(1,2, figsize=(15,6)) gp.plot.heatmap(np.array(B), ax=axs[0], title="Block Matrix", xticklabels=["L", "R"], yticklabels=["L", "R"], cbar=False, annot=True) gp.plot.heatmap(P, ax=axs[1], inner_hier_labels=zvec, title="Probability Matrix") ``` --- ### Defining the Parameters of an SBM <center>  </center> --- ### Simulating an SBM ```python # simulate an adjacency matrix A = gp.simulations.sbm(ns, B, directed=False, loops=False) # ...and plot it fig, axs = plt.subplots(1,2, figsize=(15,6)) gp.plot.heatmap(A, ax=axs[0], inner_hier_labels=zvec, title="$SBM_{70}(\\vec z, B)$ simulation") gp.plot.heatmap(P, ax=axs[1], inner_hier_labels=zvec, title="Probability Matrix") ``` --- #### The community structure is totally obvious... <center>  </center> --- ### ... so why do we care? - the community structure is .ye[only] obvious for an ordering of the nodes in community order - we don't know community ahead of time ```python import numpy as np # generate a reordering of the n nodes vtx_perm = np.random.choice(np.sum(ns), size=np.sum(ns), replace=False) Aperm = A[tuple([vtx_perm])] [:,vtx_perm] ``` ```python gp.plot.heatmap(Aperm, title="$SBM_{70}(\\vec z, B)$, random node order") ``` --- ### ... so why do we care?  --- name:commdetect ### Outline - [Background](#back) - [SBMs](#sbm) - Community Detection - [Model Selection](#modselect) --- ### How do we "guess" community structure? - Community detection: identifying unknown community labels - Idea: nodes sharing a community tend to be connected to similar nodes - why? block matrix - probability matrix is "low rank": only need one vector per community - if network has $K$ communities, $P$ is rank-$K$ --- ### What do we mean by "low-rank"? - probability matrix only has two unique row/column vectors  --- ### How do we "guess" low-rank structure? - when we have a low-rank structure, a way to deduce this structure is a spectral embedding - PCA but on a "similarity" matrix - in what sense were the nodes of the same community similar? --  -- - probability, so we use the probability matrix --- ### How do we "guess" low-rank structure? ```python # diagonal augmentation of the probability matrix Paug = gp.utils.augment_diagonal(P) # embed using spectral embedding U, s, _ = np.linalg.svd(Paug) Pembed = U[:,0:2] @ np.diag(s[0:2]) ``` ```python import seaborn as sns fig, ax = plt.subplots(1,1, figsize=(6,6)) sns.heatmap(Pembed, ax=ax) ``` --- ### How do we "guess" low-rank structure? 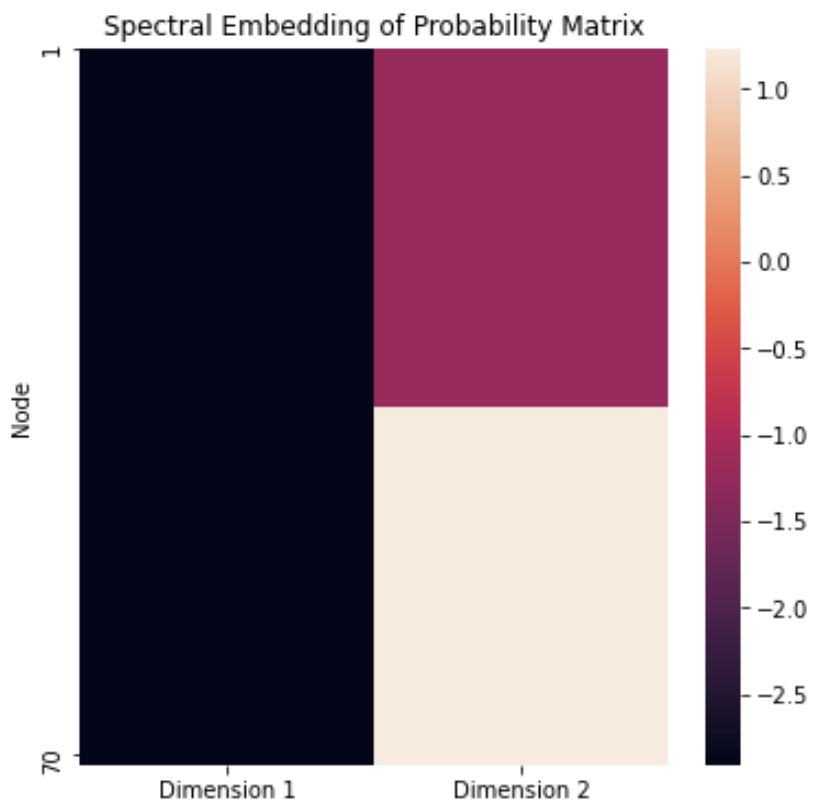 --- ### Finding a surrogate for the probability matrix - Problem: we don't know the probability matrix ahead of time - we never know the "true" underlying statistical model - what can we use? -- - it turns out that the adjacency matrix works well as a surrogate - ASE: .ye[A]djacency .ye[S]pectral .ye[E]mbedding ```python ase = gp.embed.AdjacencySpectralEmbed() Aembed = ase.fit_transform(A) ``` --- ### How do we actually deduce community labels? ```python fig, ax = plt.subplots(1,1, figsize=(6,6)) sns.heatmap(Aembed, ax=ax) ``` 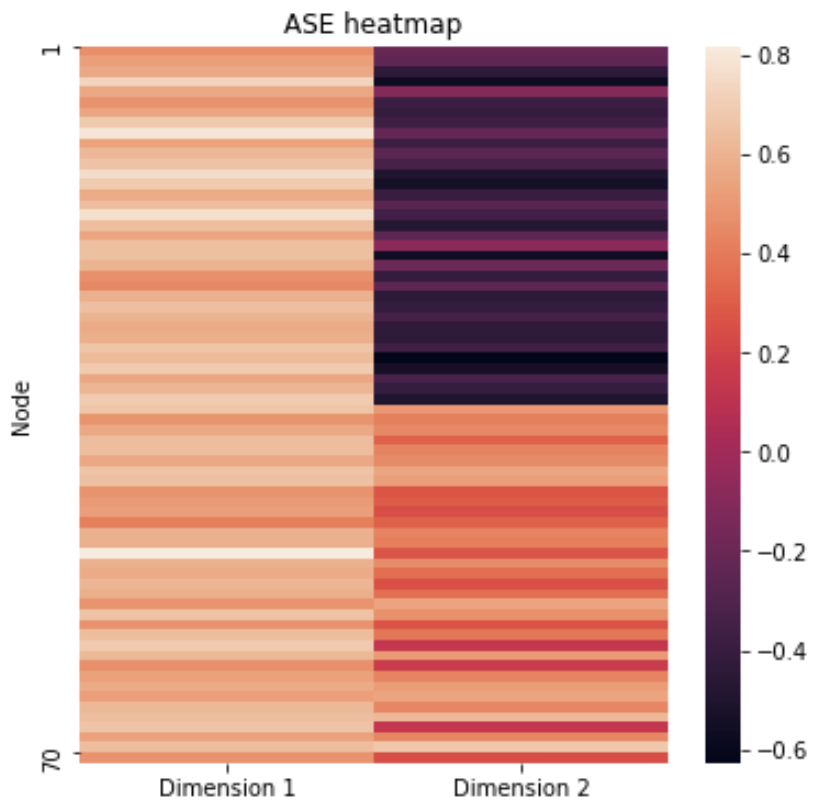 --- ### How do we actually deduce community labels? ```python gp.plot.pairplot(Aembed, labels=zvec, legend_name="Community (Unknown)", title="ASE Pairplot") ``` 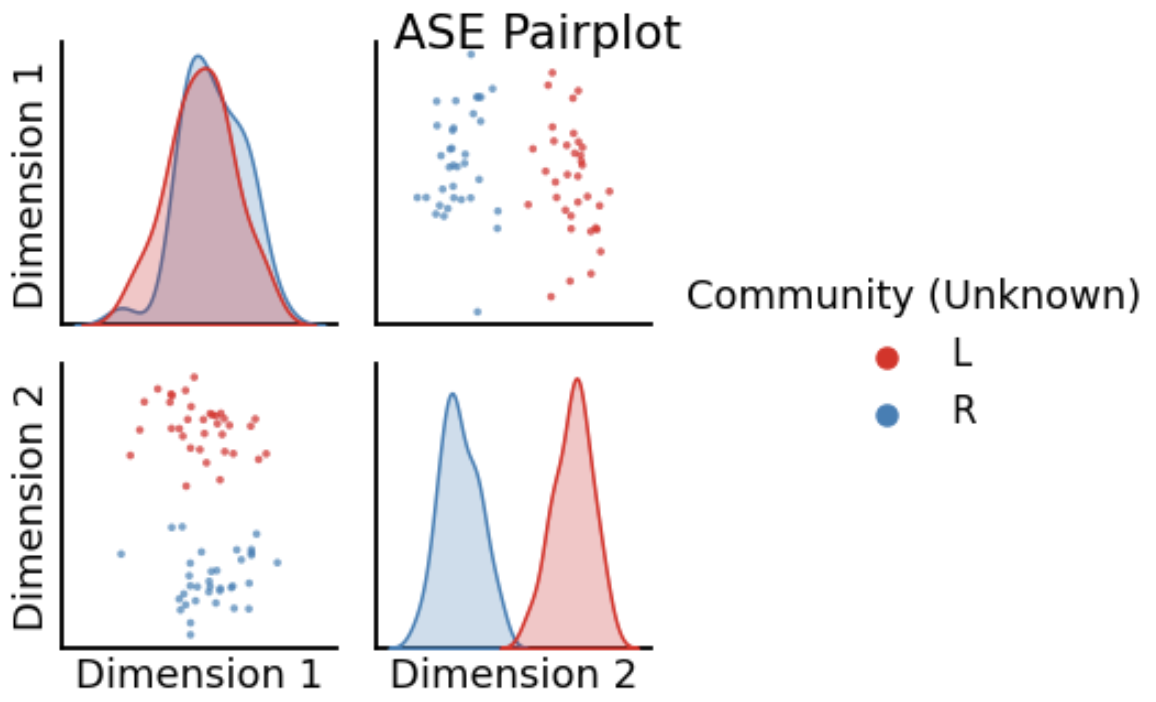 --- ### Community Detection - take the ASE and cluster it - number of clusters = number of communities you expect -- - evaluation: choose a metric which is high for "good" clusterings, and "low" for bad clusterings - Kmeans: ARI (.ye[A]djusted .ye[R]and .ye[I]ndex) ```python from sklearn.cluster import KMeans from sklearn.metrics import adjusted_rand_score labels_kmeans = KMeans(n_clusters = 2).fit_predict(Aembed) ari_kmeans = adjusted_rand_score(labels_kmeans, zvec) ``` --- ### Evaluation of ASE ```python gp.plot.pairplot(Aembed, labels=labels_kmeans, legend_name="Predicted", title="ASE Pairplot (ARI={:.2f})".format(ari_kmeans)) ``` 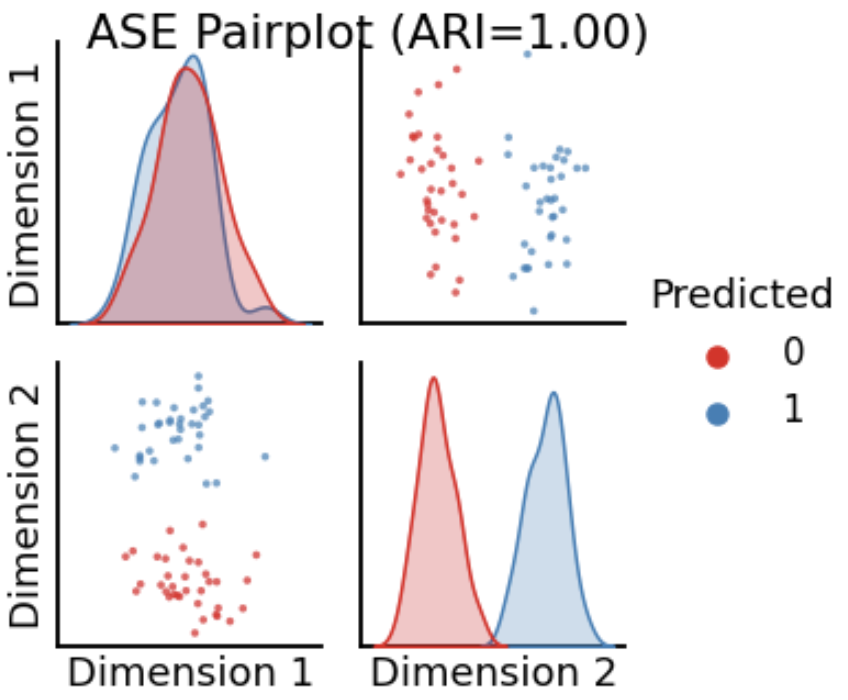 --- ### Evaluation of ASE - clustering is unsupervised: which cluster is Left vs. Right for examining error rate? - `gp.utils.remap_labels()`: "rename" (0/1 to Left/Right) labels of the unsupervised predictions - as many samples as possible have the same label in unsupervised predictions and the true labels ```python # remap with graspologic labels_kmeans_remap = gp.utils.remap_labels(zvec, labels_kmeans.astype(str)) # errors are where the prediction != the true label error = zvec != labels_kmeans_remap # error rate is the frequency of making an error er_rt = np.mean(error) palette = {'Right':(0,0.7,0.2), 'Wrong':(0.8,0.1,0.1)} # initialize numpy array for each node error_label = np.array(len(zvec)*['Right']) # add label 'Wrong' for each error that is made error_label[error] = '›' ``` --- ### Evaluation of ASE ```python gp.plot.pairplot(Aembed, labels=labels_kmeans_remap, legend_name="Predicted", title="ASE Pairplot (ARI={:.2f})".format(ari_kmeans)) ```  --- ### Evaluation of ASE ```python gp.plot.pairplot(Aembed, labels=error_label, legend_name="Prediction successful?", title="ASE Pairplot (Error={:.2f})".format(er_rt), palette=palette) ```  --- ### How do we decide the number of communities? - If we don't know the number of communities ahead of time, what do we do? -- - use a performance metric! - a good choice tends to be the Silhouette score - clusterings with high Silhouette $\Rightarrow$ better - clusterings with low Silhouette $\Rightarrow$ worse -- - repeat clustering for "plausible" numbers of clusters, and pick the one where Silhouette is highest ```python from pandas import DataFrame as df comm_select = gp.cluster.KMeansCluster(max_clusters=10) comm_select = comm_select.fit(Aembed) nclusters = range(2, 10) silhouette = comm_select.silhouette_ silhouette_df = df({"Number of Clusters": nclusters, "Silhouette Score": silhouette}) ``` --- ### How do we decide the number of communities? ```python fig, ax = plt.subplots(1,1,figsize=(7, 4)) sns.lineplot(data=silhouette_df,ax=ax, x="Number of Clusters", y="Silhouette Score"); ax.set_title("Silhouette Analysis of KMeans Clusterings") ax.set_ylim([-1, 1]) ```  --- name:modselect ### Outline - [Background](#back) - [SBMs](#sbm) - [Community Detection](#commdetect) - Model Selection --- --- ### Links - [Graspologic python package](https://github.com/microsoft/graspologic) - [Graspologic book repository](https://github.com/neurodata/graph-stats-book) - [Graspologic book](http://docs.neurodata.io/graph-stats-book/coverpage.html) - Questions?